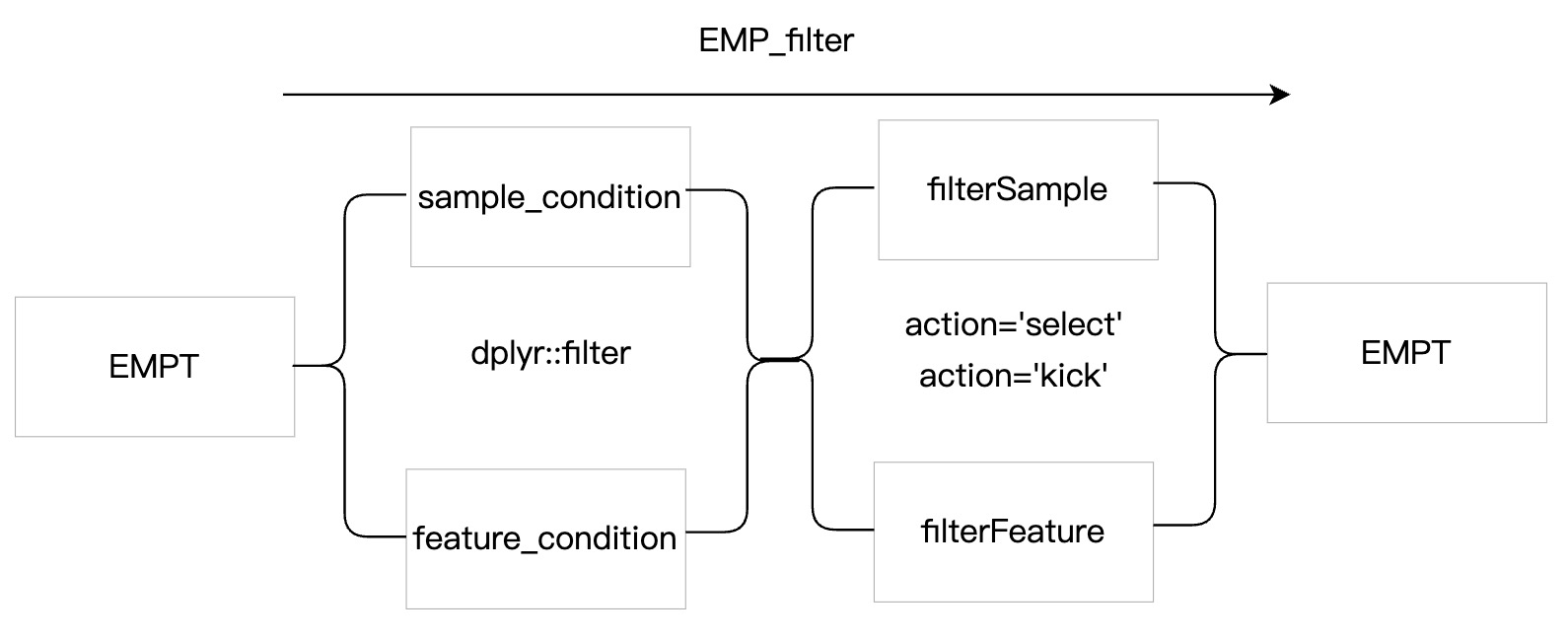
7.1 EMP_filter
The function of module EMP_filter is very powerful, which can not only filter features and samples according to rowdata and coldata but also filter them according to data analysis results. To facilitate the user's understanding, the following describes the basic parameters of the module EMP_filter:
- obj: Specify the object to be analyzed, either
MAEorEMPT. - experiment: Specify the name of the project to be analyzed (character).
- sample_condition: Specify the threshold conditions for selecting samples.
- feature_condition: Specify the threshold conditions for selecting features.
- filterSample: Specify the names of the samples to be filtered.
- filterFeature: Specify the names of the features to be filtered.
- action: Used in conjunction with parameters
filterSampleandfilterFeature, set tokickorselect. - keep_result: If the input is TRUE, it means to keep all analysis results, regardless of how samples and features change. If the input is a name, it means to keep the corresponding analysis results.
- show_info: Specify the display method of the output results.
① It is necessary to understand the filtering logic in the module
EMP_filter. First, samples and features are filtered based on the conditions specified by the parameters sample_condition and feature_condition. Then, samples and features are selected or kicked based on the parameters filterSample and filterFeature. ② If
EMP_filter operation does not meet the filtering requirements in once time, the EMP_filter process can be performed multiple times.
7.1.1 Filter the data across all the experiments in MAE
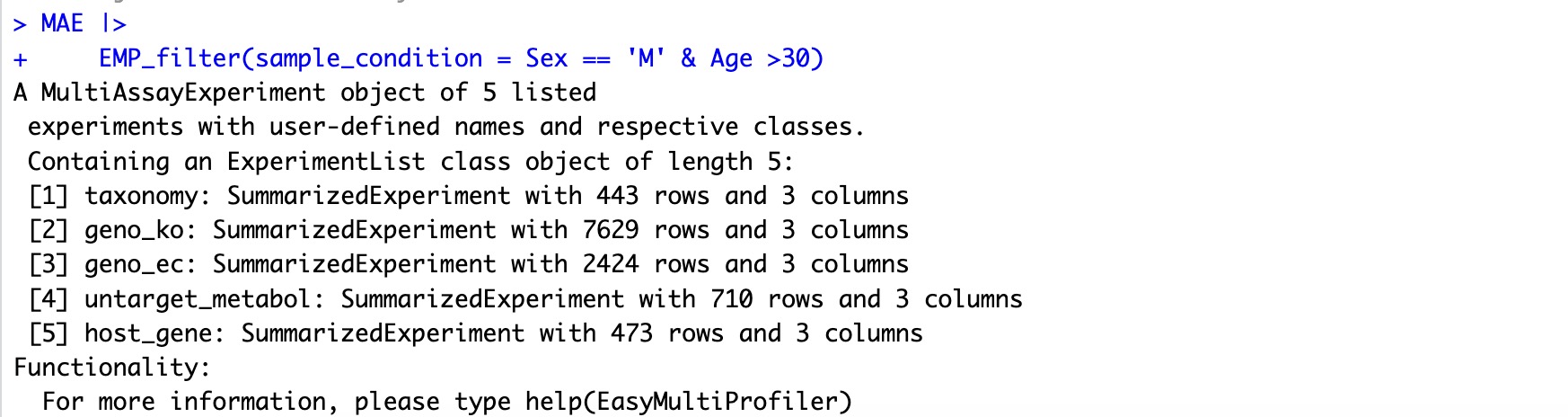
🏷️Example 1:Screen male subjects older than 30 years from all omics projects.
MAE |>
EMP_filter(sample_condition = Sex == 'M' & Age >30)
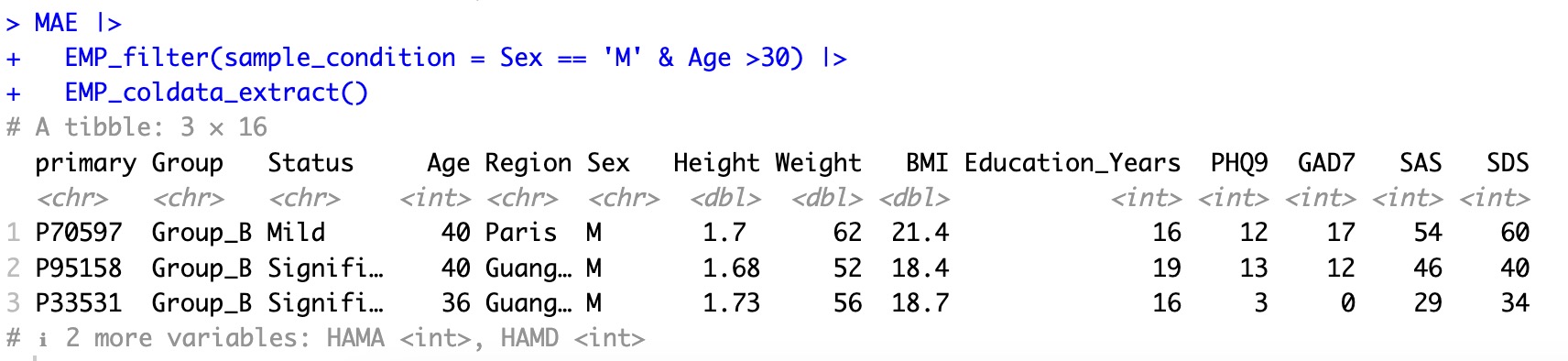
After filtering, only the samples that meet the conditions that exist in the MAE object, and other analysis modules can be directly used for downstream analysis. For example, users can use the module EMP_coldata_extract to observe the filtered sample.
MAE |>
EMP_filter(sample_condition = Sex == 'M' & Age >30) |>
EMP_coldata_extract()
🏷️Example 2:Select the samples without missing values in the sample-related data.
The module
EMP_filter inherits the filter syntax from the dplyr package, so the tidy syntax for filtering is avaviable.
MAE |>
EMP_filter(sample_condition = if_all(everything(),~ !is.na(.)))
🏷️Example 3:Select the samples whose at least one of the PHQ-9 or GAD-7 scores is greater than 5.
MAE |>
EMP_filter(sample_condition = if_any(c(PHQ9,GAD7),~. > 5))
7.1.2 Filter samples and features in single omics data
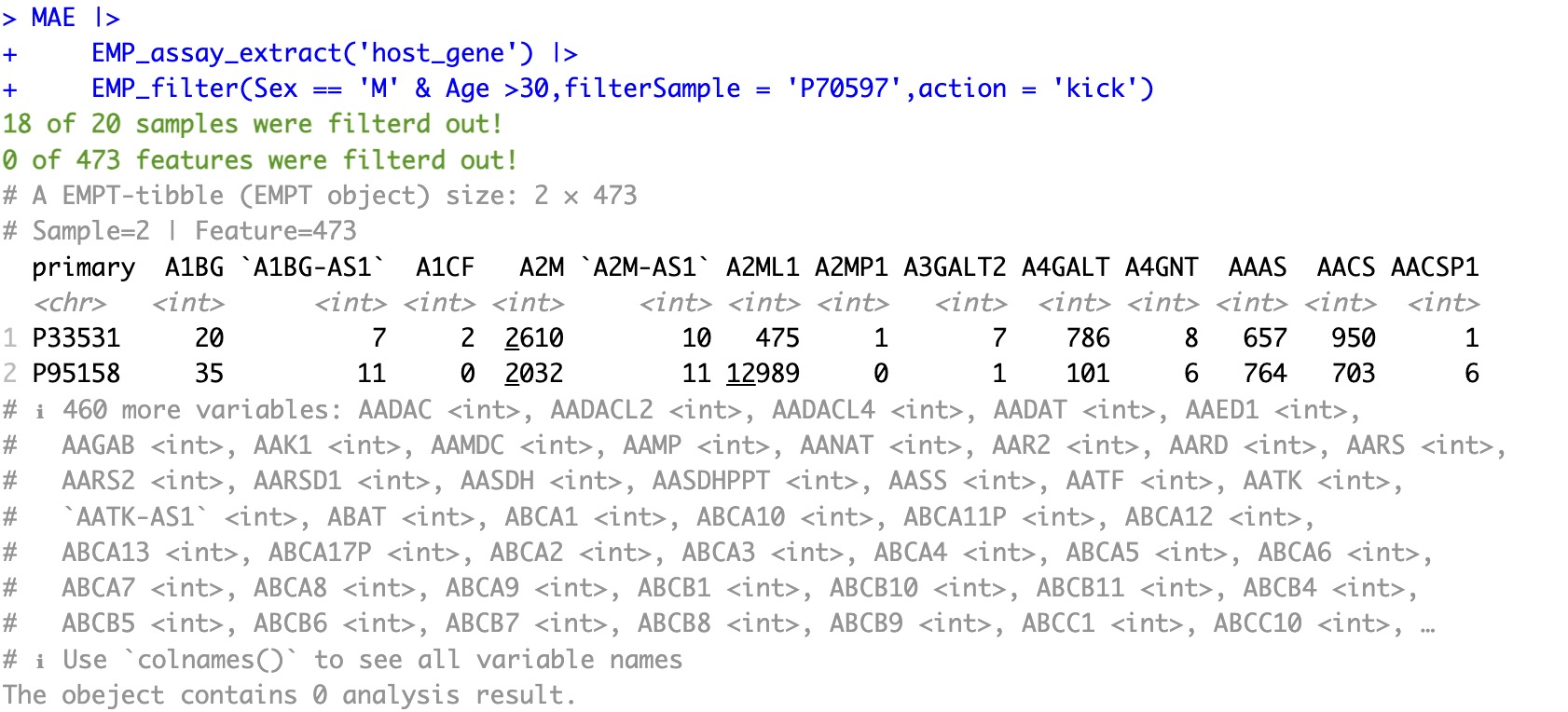
🏷️Example 1:Filter samples based on raw data.
Select male samples over the age of 30 from the data and exclude the sample P70597.
The parameter
action is used only for the parameters filterSample and filterFeature.MAE |>
EMP_assay_extract('host_gene') |>
EMP_filter(Sex == 'M' & Age >30,filterSample = 'P70597',action = 'kick')
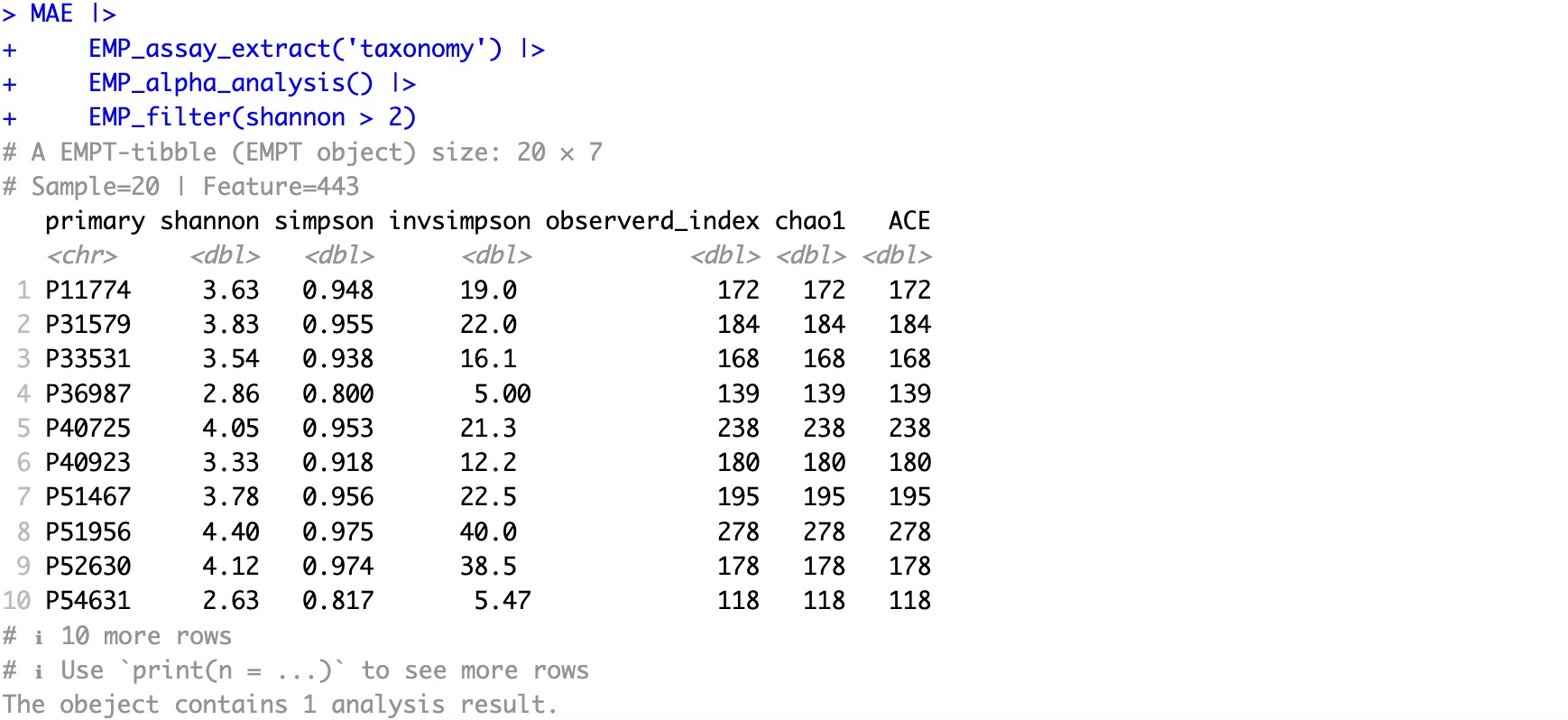
🏷️Example 2:Filter samples based on previous alpha analysis result. Select samples with the Shannon value greater than 2.
MAE |>
EMP_assay_extract('taxonomy') |>
EMP_alpha_analysis() |>
EMP_filter(shannon > 2)
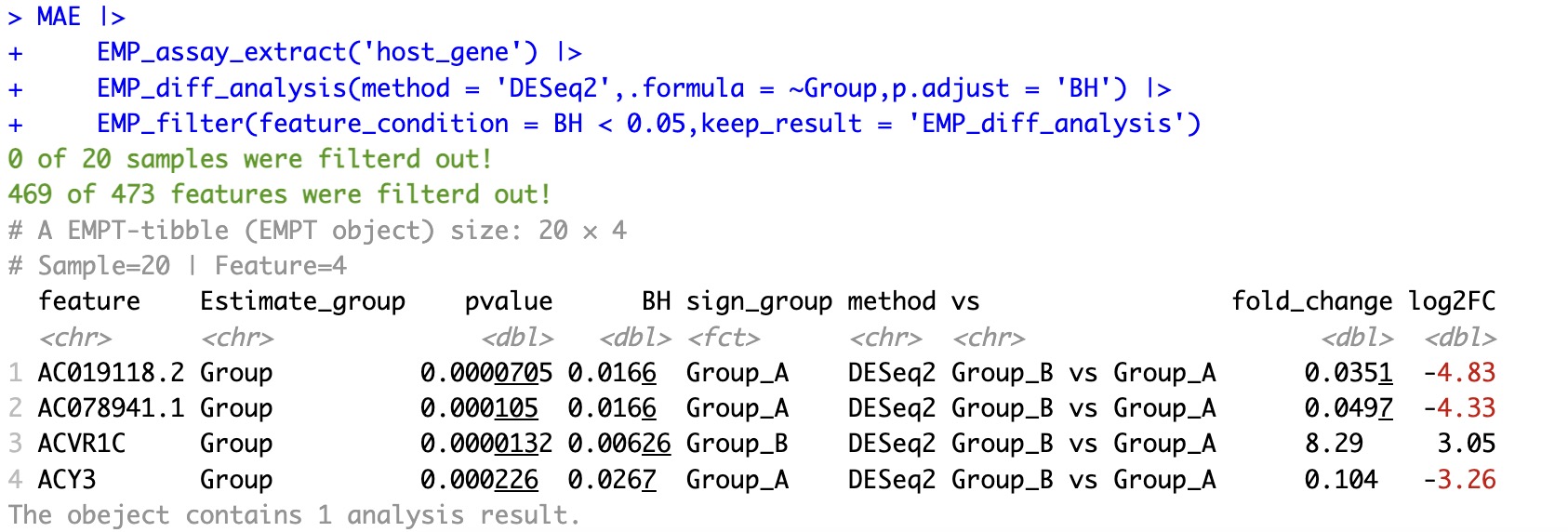
🏷️Example 3:Filter samples based on previous difference analysis result. Select feature with the adjust P value less than 0.05.
①In the DESeq2 algorithm, the differential analysis results will change when features and samples are modified. Therefore, if you need to retain the original differential analysis results, the parameter
keep_result is required.②Traditional statistical tests, such as t.test or wilcox.test, are not affected by changes in features, so there is no need for the parameter
keep_result to retain these results.
MAE |>
EMP_assay_extract('host_gene') |>
EMP_diff_analysis(method = 'DESeq2',.formula = ~Group,p.adjust = 'BH') |>
EMP_filter(feature_condition = BH < 0.05,keep_result = 'EMP_diff_analysis')
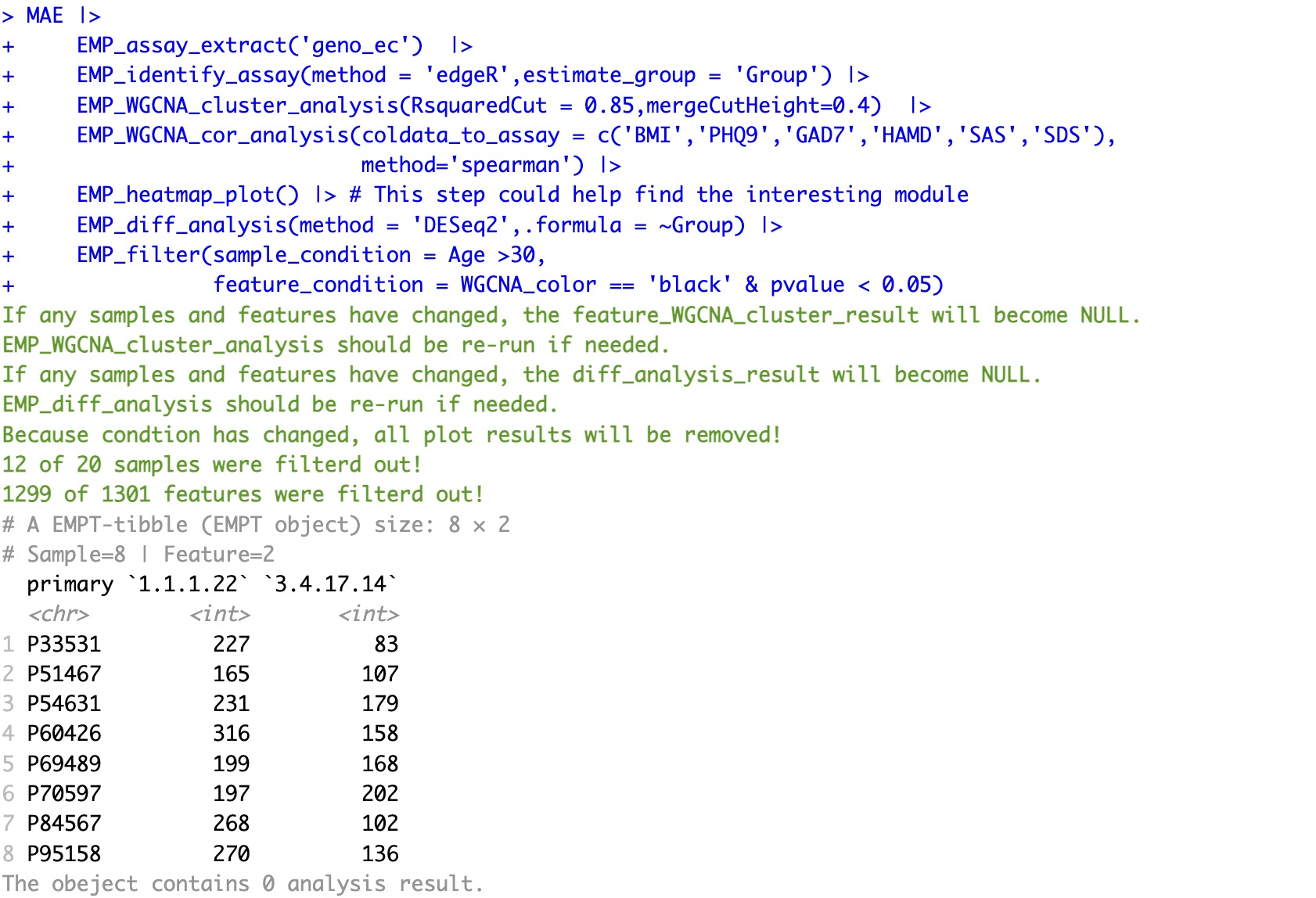
🏷️Example 4: Filter samples and features in multiple conditions and times.
Extract the assay of geno_ec. Perform WGCNA analysis followed by DESeq2 difference analysis, and then filter the data based on the following criteria:
- Samples older than 30 years;
- Core genes identified by edgeR;
- Genes from the WGCNA analysis that are associated significantly with BMI;
- Genes with a p-value less than 0.05 in the difference analysis.
MAE |>
EMP_assay_extract('geno_ec') |>
EMP_identify_assay(method = 'edgeR',estimate_group = 'Group') |>
EMP_WGCNA_cluster_analysis(RsquaredCut = 0.85,mergeCutHeight=0.4) |>
EMP_WGCNA_cor_analysis(coldata_to_assay = c('BMI','PHQ9','GAD7','HAMD','SAS','SDS'),
method='spearman') |>
EMP_heatmap_plot() |> # This step can help find the interesting module
EMP_diff_analysis(method = 'DESeq2',.formula = ~Group) |>
EMP_filter(sample_condition = Age >30,
feature_condition = WGCNA_color == 'black' & pvalue < 0.05)
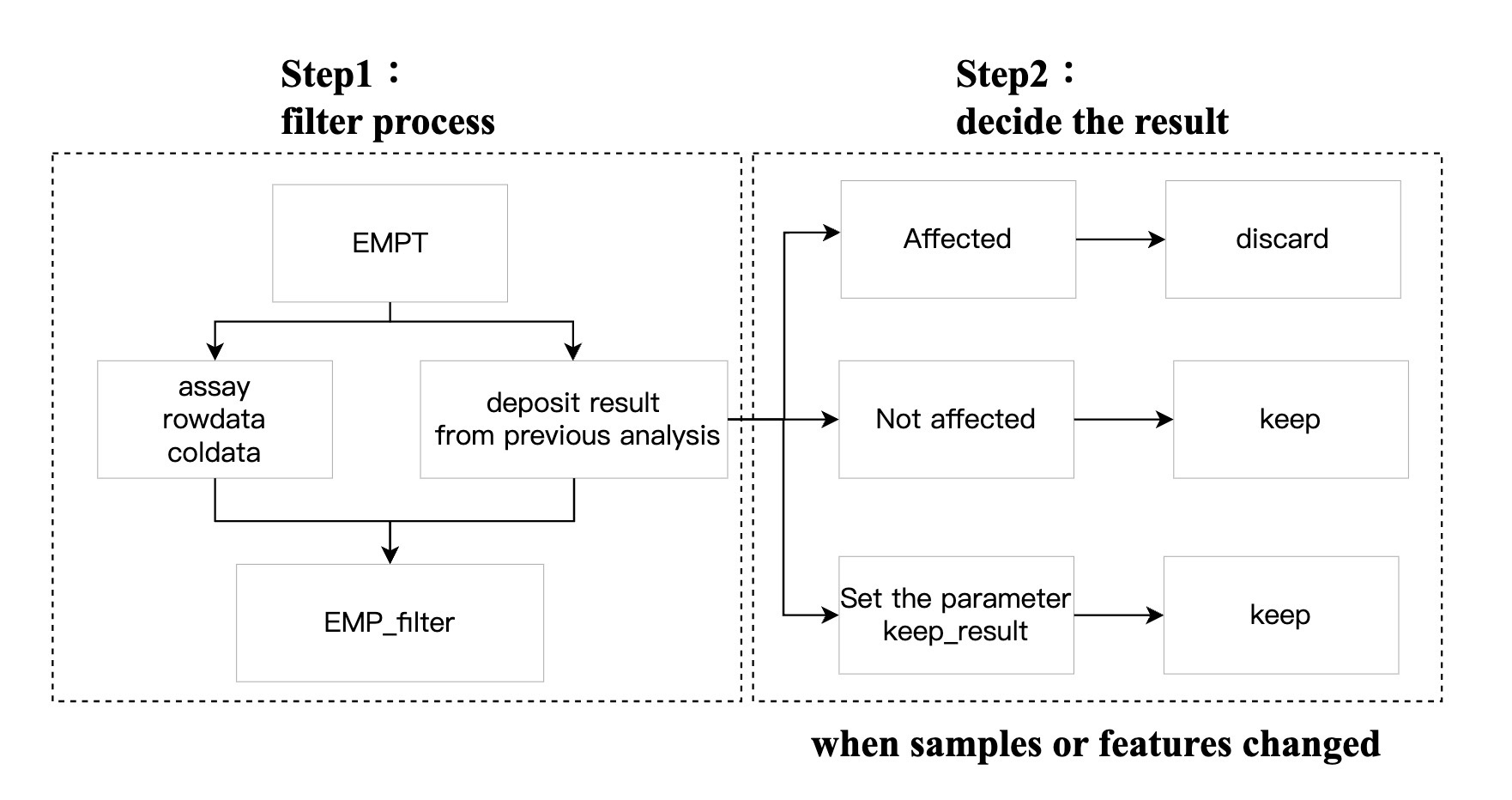
7.1.3 Regarding the removal of data analysis results after filtering
In the analysis workflow in the EasyMultiProfiler package, the analysis results are automatically stored in objects for the module EMP_filter. However, once the screening is complete, the previously stored results may no longer be correct and are therefore automatically cleared away (e.g., the results of the difference analysis and alpha diversity analysis are stored in the container, and then some samples are excluded based on the BMI value). At this stage, the existing difference analysis results are automatically cleared because they are no longer right; The alpha diversity results are unaffected by the sample change and will continue to be stored in the object for the next screening). The EasyMultiProfiler package already automates this process and will alert the user in red text that the action has been performed.
Examples that are easy to misunderstand:
🏷️Example : Perform the differential analysis of EC 1.1.1.1 using DESeq2 algorithm
MAE |>
EMP_assay_extract('geno_ec') |>
EMP_diff_analysis(method = 'DESeq2',.formula = ~Group)
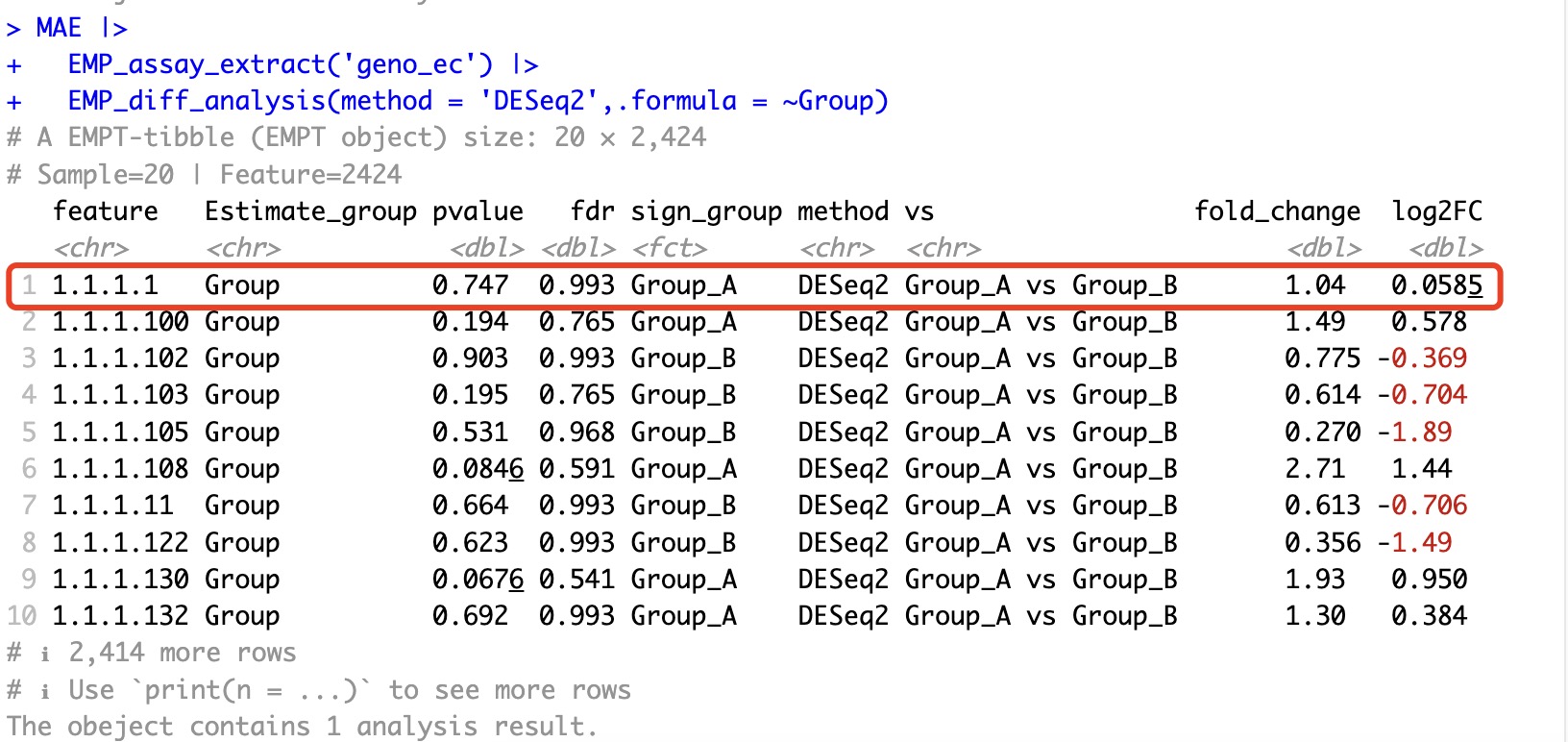
After removing only some of the other features and performing DESeq2 differential analysis again, we find that the results for 1.1.1.1 have changed. This is due to the inherent characteristics of the DESeq2 algorithm. Similar situations also occur with algorithms like edgeR and limma.
MAE |>
EMP_assay_extract('geno_ec',pattern = '1.1.1.1',pattern_ref = 'feature') |>
EMP_diff_analysis(method = 'DESeq2',.formula = ~Group)
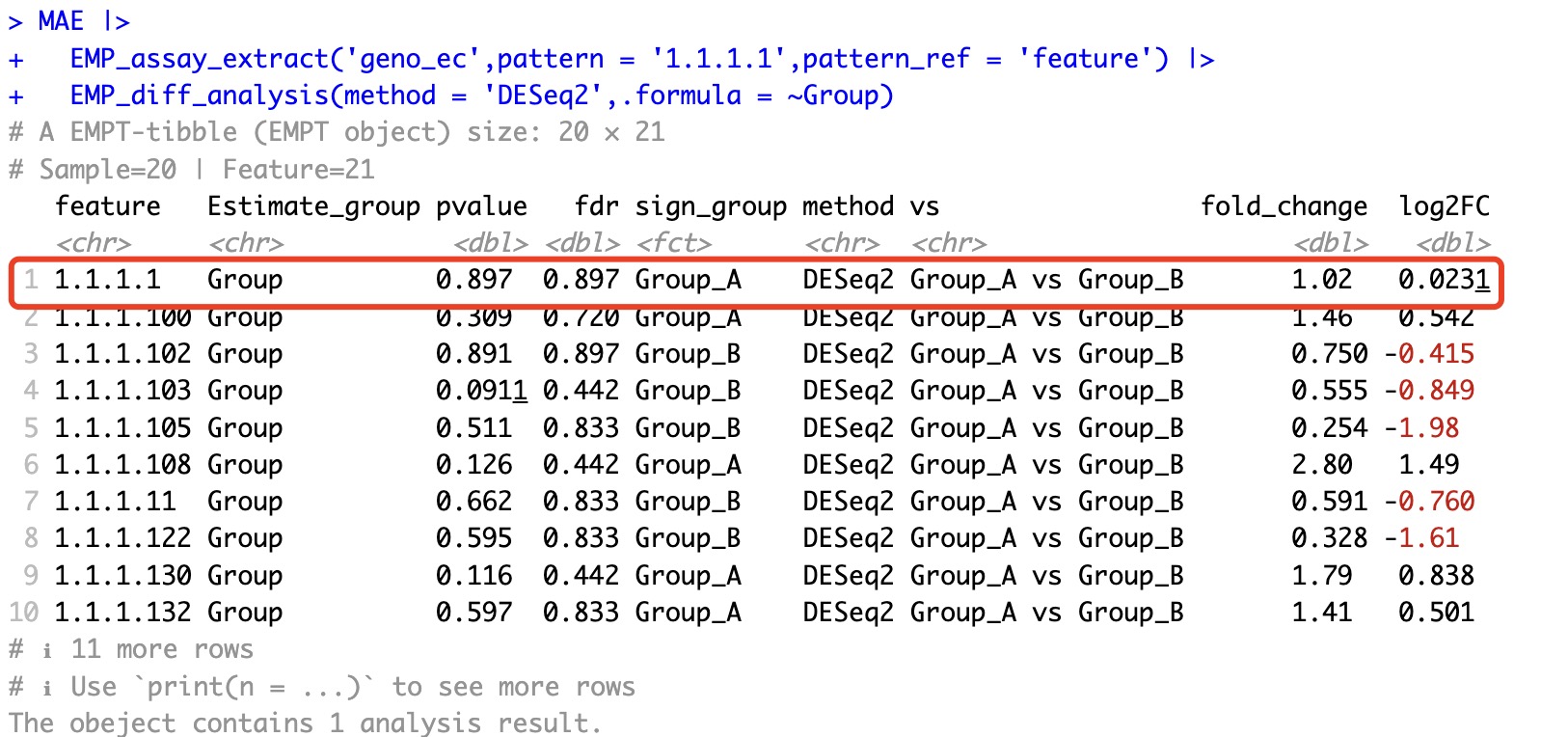
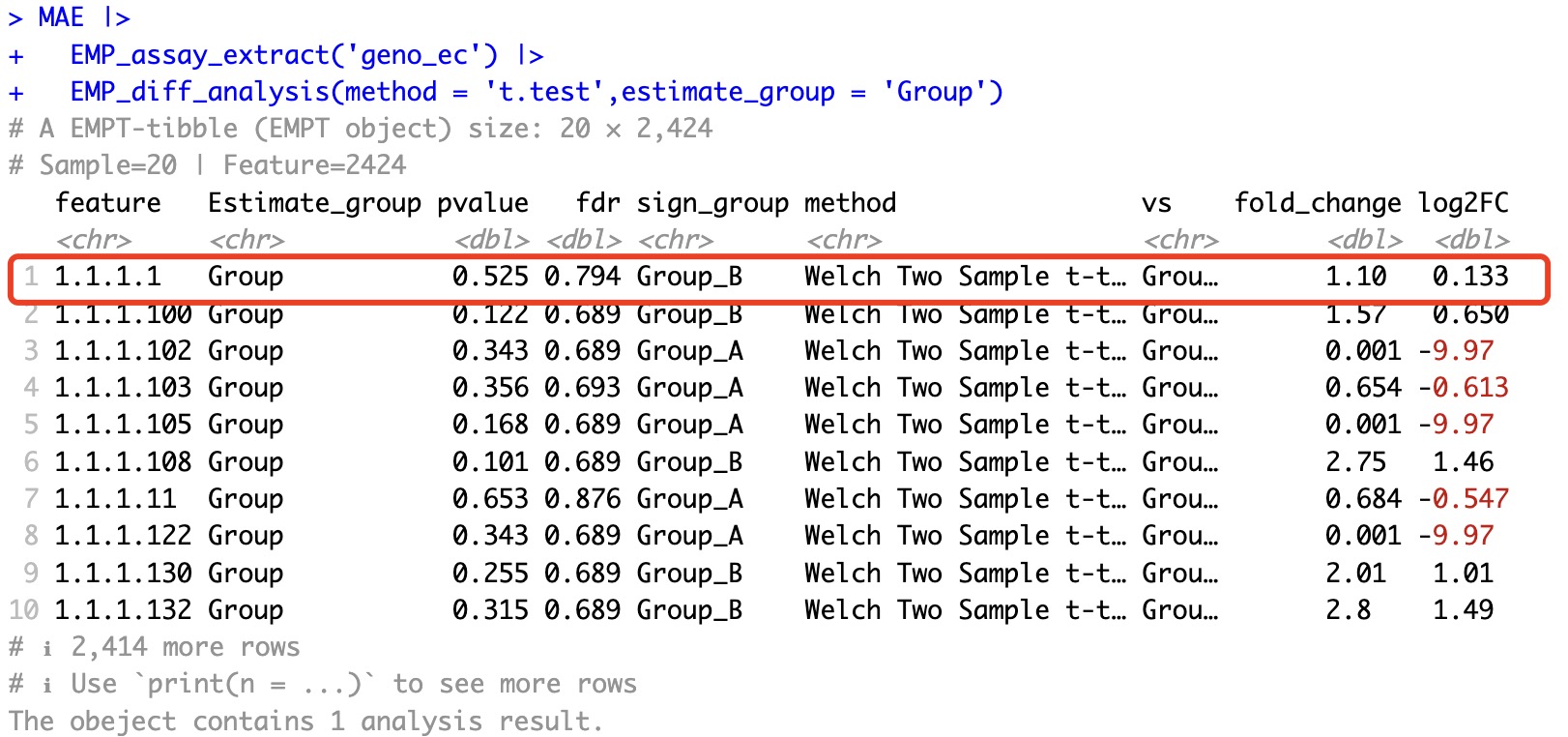
However, such situations do not occur in traditional statistical tests.
MAE |>
EMP_assay_extract('geno_ec') |>
EMP_diff_analysis(method = 't.test',estimate_group = 'Group')
MAE |>
EMP_assay_extract('geno_ec',pattern = '1.1.1.1',pattern_ref = 'feature') |>
EMP_diff_analysis(method = 't.test',estimate_group = 'Group')
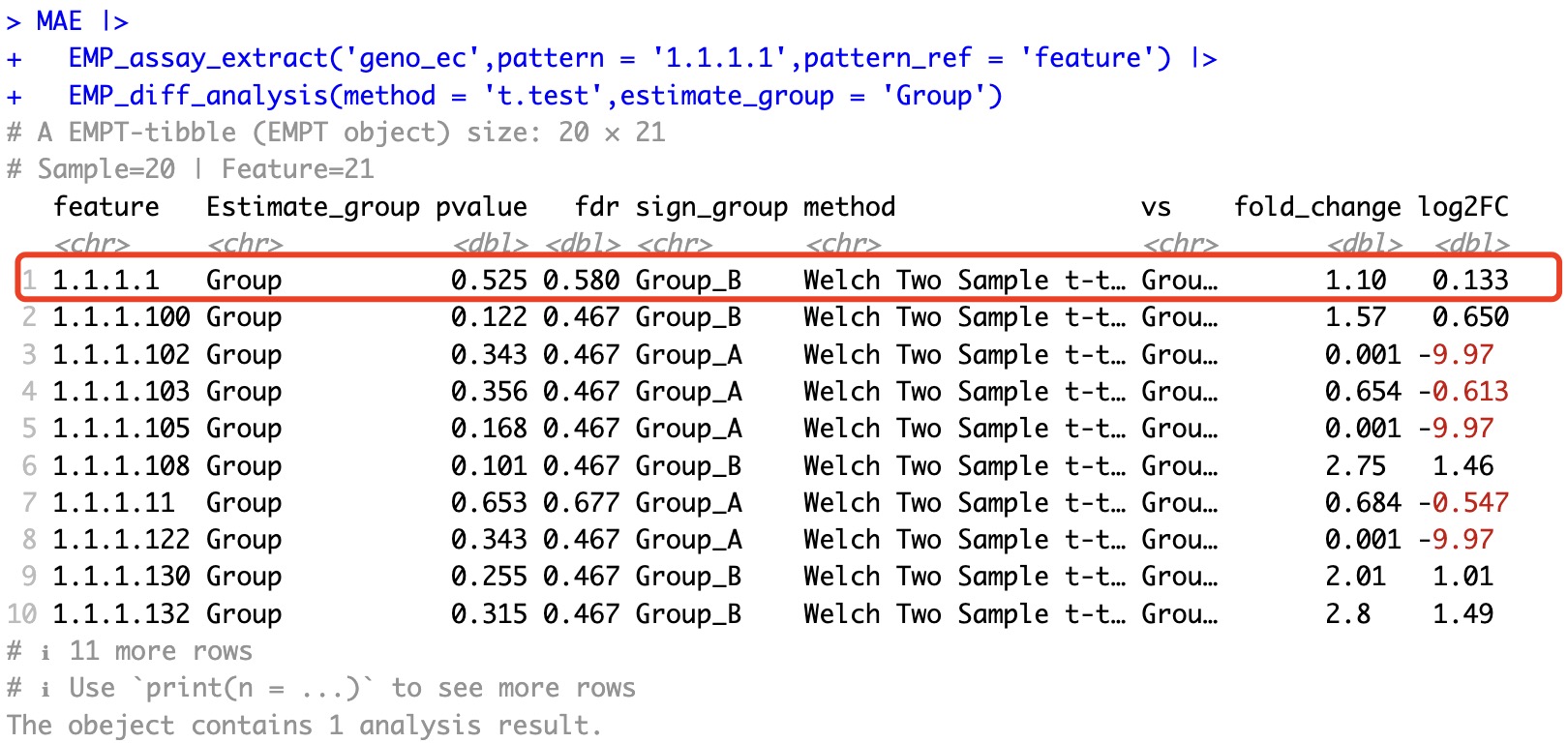
Therefore, when the number of features changes, EMP_filter will retain the differential results from traditional statistics while discarding the results from DESeq2, edgeR, and limma algorithms. For example, if we filter for features with a p-value less than 0.05 after DESeq2 differential analysis, the reduction in the number of features will trigger the removal action. As a result, after filtering, the differential analysis results will be discarded, and the output will be the assay matrix. If you want to retain the corresponding results, you can use the keep_result parameter to preserve the results from the analysis.
MAE |>
EMP_assay_extract('geno_ec') |>
EMP_diff_analysis(method = 'DESeq2',.formula = ~Group) |>
EMP_filter(feature_condition = pvalue < 0.05)
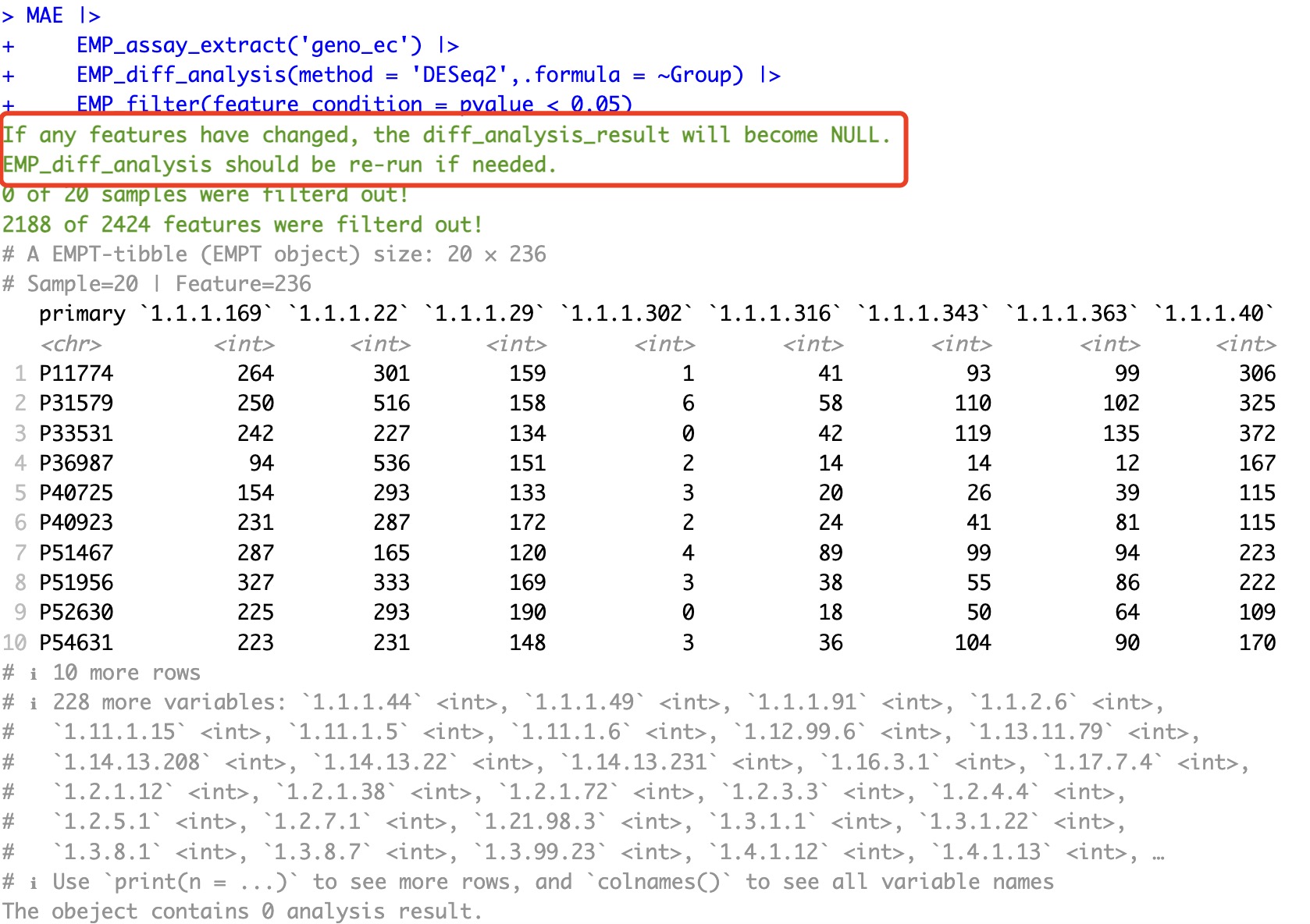
If you want to retain the corresponding results, you can use the keep_result parameter to preserve the results from the analysis.
MAE |>
EMP_assay_extract('geno_ec') |>
EMP_diff_analysis(method = 'DESeq2',.formula = ~Group) |>
EMP_filter(feature_condition = pvalue < 0.05,keep_result = 'EMP_diff_analysis')
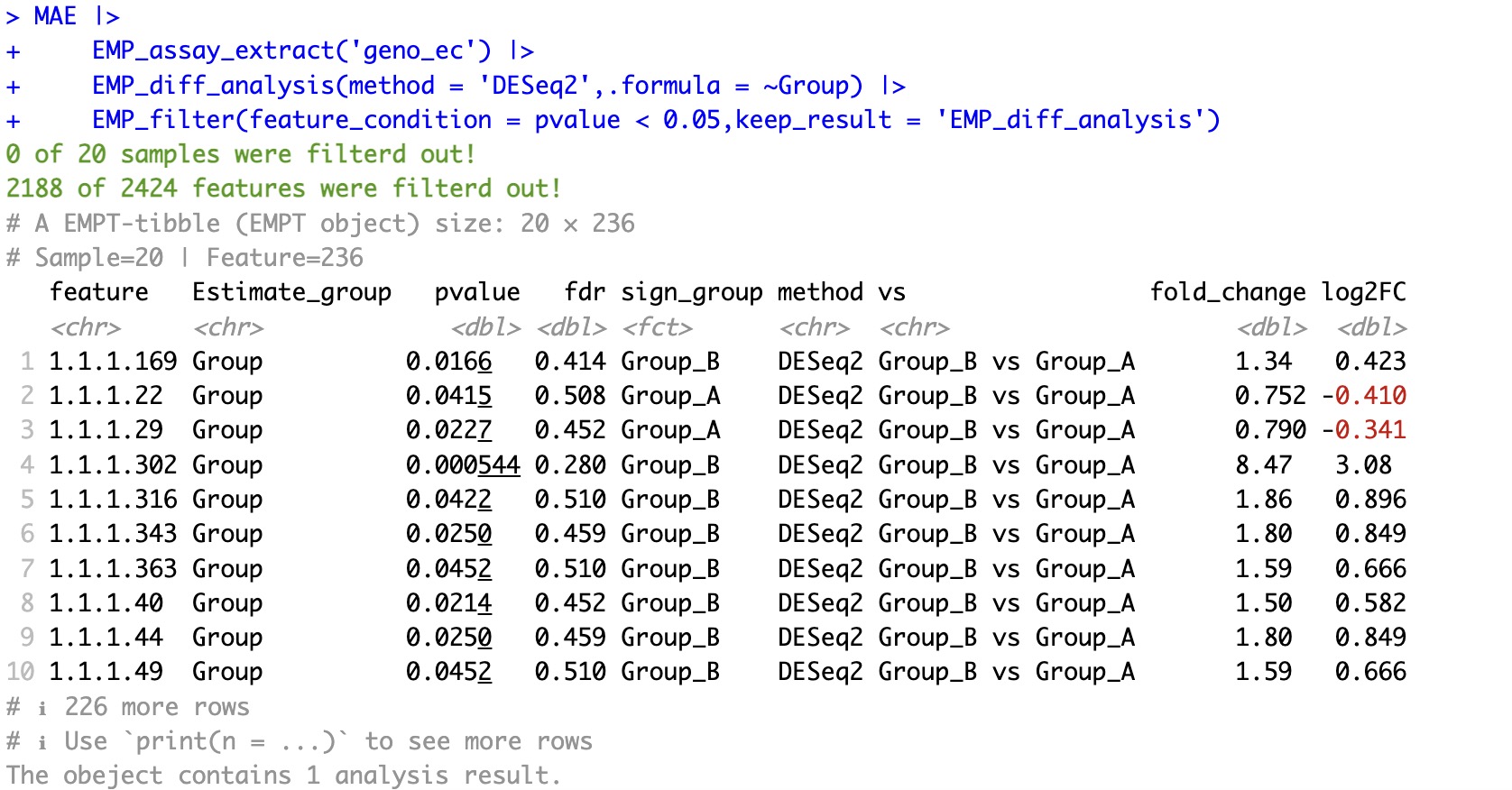
7.1.4 Regarding the display of filtered data
In the analysis workflow of the EasyMultiProfiler package, after filtering with the module EMP_filter, the output will by default retain the display results from the previous analysis module. For example, if the previous module performed alpha diversity calculations, and EMP_filter is used to filter based on the Age>30 condition, the EMP_filter output will continue to display the filtered results from the previous module. If the result is cleared due to changes in samples or features (as detailed in section 7.1.3), the output will instead reflect the current state of the dataset assay.
7.1.5 Regarding filtered Data Extraction
In the module EMP_filter, the parameter action is used only to filter with parameters filterSample and filterFeature. If users want to extract the assay data after filtering, it is a good idea to use the module EMP_assay_extract after the module EMP_filter.
🏷️Example:Extract assay after filtering.
MAE |>
EMP_assay_extract('host_gene') |>
EMP_filter(Sex == 'M' & Age >30,filterSample = 'P70597',action = 'kick') |>
EMP_assay_extract(action = 'get')
7.1.6 Variable referencing in filter operations
The EMP_filter module inherits its syntax from dplyr's filter function. When you introduce some indirection, i.e. when you want to get the data-variable from an env-variable instead of directly typing the data-variable’s name, you need to embrace the argument by surrounding it in doubled braces, like .
Always use this proper indirection syntax to avoid incorrect filtering results.
🏷️Example:
target_status = c("Mild","No")
MAE |>
EMP_filter(Status %in% target_status) ## Wrong usage!!
MAE |>
EMP_filter(Status %in% {{target_status}}) ## Correct usage!!